The E.C. 6 class of ligase enzymes includes synthases and synthetases. They participate in synthetic reactions and catalyze the combining of two molecules while simultaneously breaking the diphosphate link in ATP or another comparable triphosphate.
In contrast to synthetases, which use ATP as a source of energy during the synthesis of biological compounds, synthases are any ligases that catalyze the synthesis of biological compounds without using ATP as an energy source.
In this article, you’ll get to know exactly the difference between Synthase and Synthetase.
What Is Synthase?
A synthase is an enzyme that catalyzes the synthesis process in biochemistry. Keep in mind that the biological nomenclature initially distinguished between synthetases and synthases.
In accordance with the original definition, synthetases utilize nucleoside triphosphates (such as ATP, GTP, CTP, TTP, and UTP) as a source of energy while synthases do not.
Nevertheless, according to the Joint Commission on Biochemical Nomenclature (JCBN), “synthase” can be used to refer to any enzyme that catalyzes synthesis (regardless of whether it employs nucleoside triphosphates), but “synthetase” should only be used to refer to “ligase.”
Here is a list of examples of different types of synthase:
- ATP synthase
- Citrate synthase
- Tryptophan synthase
- Pseudouridine synthase
- Fatty acid synthase
- Cellulose synthase (UDP-forming)
- Cellulose synthase (GDP-forming)
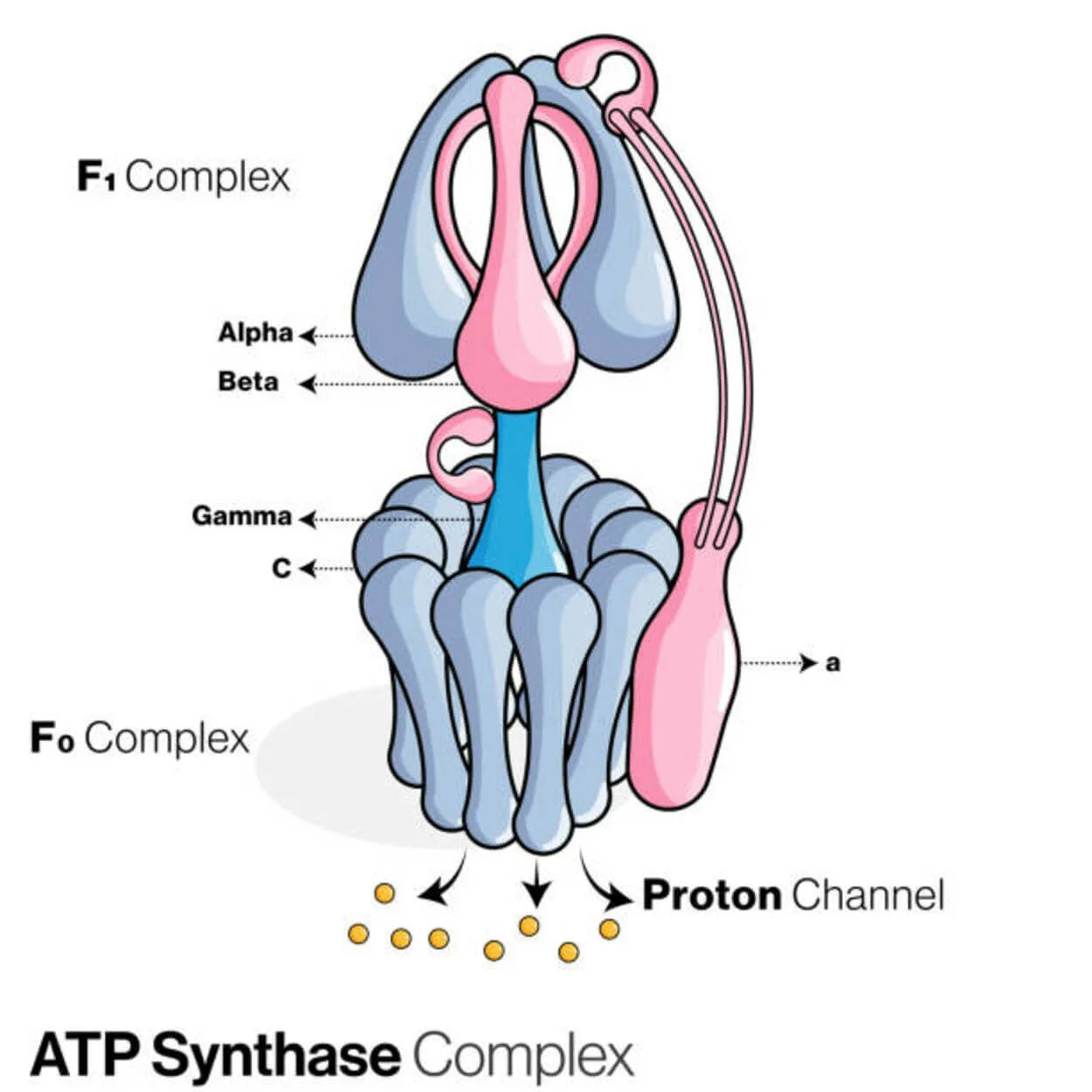
ATP Synthase
Adenosine diphosphate (ADP) and inorganic phosphate are used to create the energy storage molecule adenosine triphosphate (ATP) by a protein called ATP synthase (Pi).
It’s categorized as a ligase since it modifies ADP by forming a P-O link (phosphodiester bond). A molecular device called ATP synthase.
Energy-wise, the production of ATP from ADP and Pi is undesirable, and the process would typically go the other way.
A proton (H+) concentration gradient across the inner mitochondrial membrane in eukaryotes or the plasma membrane in bacteria drives this reaction forward by coupling ATP synthesis during cellular respiration to the gradient.
In plants, ATP synthase uses a proton gradient formed in the thylakoid lumen across the thylakoid membrane and into the chloroplast stroma to produce ATP during photosynthesis.
For an ATPase, eukaryotic ATP synthases are F-ATPases that function “in reverse.” This kind is primarily discussed in this article. The FO and F1 subunits of an F-ATPase have a rotational motor mechanism that enables ATP synthesis.
Citrate Synthase
Nearly all living cells contain the enzyme citrate synthase, which serves as a pacemaker in the first step of the citric acid cycle and is designated E.C. 2.3.3.1 (previously 4.1.3.7). (or Krebs cycle).
Citrate synthase is located in the mitochondrial matrix of eukaryotic cells, although nuclear DNA, not mitochondrial DNA, encodes it.
It’s created in the cytoplasm by cytoplasmic ribosomes and subsequently moved into the mitochondrial matrix.
A typical quantitative enzyme marker for the existence of intact mitochondria is citrate synthase. Citrate synthase’s peak activity reveals how many mitochondria are present in skeletal muscle.
High-intensity interval training has the potential to raise maximum activity more than either endurance training or high-intensity interval training.
Acetyl coenzyme A has two-carbon acetate residue and a molecule of four-carbon oxaloacetate condenses to generate six-carbon citrate, which is produced by the condensation reaction catalyzed by citrate synthase.
Tryptophan Synthase
The final two steps in the production of tryptophan are catalyzed by the enzyme tryptophan synthase, also known as tryptophan synthetase.
Eubacteria, Archaebacteria, Protista, Fungi, and Plantae are frequent hosts to it. However, Animalia doesn’t have it. Usually, it appears as a 2 2 tetramer.
The subunits catalyze the reversible conversion of indole-3-glycerol phosphate to indole and glyceraldehyde-3-phosphate (G3P) (IGP).
In a pyridoxal phosphate (PLP) dependent process, the subunits catalyze the irreversible condensation of indole and serine to generate tryptophan.
An internal hydrophobic channel that is 25 angstroms long and located in the enzyme connects each active site to the adjacent active site.
This promotes substrate channeling, a mechanism by which indole produced at active sites diffuses directly to other active sites. Tryptophan synthase contains allosterically coupled active sites.
Eubacteria, Archaebacteria, Protista, Fungi, and Plantae are frequently discovered to include tryptophan synthase. Humans and other animals lack it.
One of the nine necessary amino acids for humans, tryptophan is one of the twenty standard amino acids. Tryptophan is therefore essential to the human diet.
It’s also known that tryptophan synthetase can use indole analogs, such as fluorinated or methylated indoles, as substrates to produce the equivalent tryptophan analogs.
Pseudouridine
The Greek letter psi- is used to abbreviate pseudouridine, an isomer of the nucleoside uridine in which the uracil is joined to the carbon atom by a carbon-carbon link rather than a nitrogen-carbon glycosidic connection. (Uracil is occasionally referred to as “pseudouracil” in this arrangement.)
The most prevalent RNA alteration in cellular RNA is pseudouridine. RNA can undergo over 100 chemically unique alterations during transcription and synthesis.
In addition to the four conventional nucleotides, these might potentially affect RNA expression post-transcriptionally and have a number of functions in the cell, including RNA translation, localization, and stability.
One of these is pseudouridine, a C5-glycoside isomer of uridine with a C-C bond between the C1 of the ribose sugar and the C5 of uracil in place of the typical C1-N1 bond present in uridine.
It has additional rotational mobility and conformational flexibility because of the C-C bond. In addition, the N1 position of pseudouridine possesses an additional hydrogen bond donor.
Pseudouridine, also called 5-ribosyluracil, is a familiar yet mysterious component of structural RNAs (transfer, ribosomal, small nuclear (snRNA), and small nucleolar). It was recently found in coding RNA as well.
It was the first to be detected, is the most prevalent, and can be found in all three evolutionary domains of life. In the yeast tRNA, pseudouridine makes about 4% of the nucleotides
Through the formation of additional hydrogen bonds with water, this base alteration is able to stabilize RNA and enhance base-stacking.
The number of pseudouridines grows with an organism’s complexity. There are 11 pseudouridines in the rRNA of Escherichia coli, 30 in the cytoplasmic rRNA of yeast, one alteration in the mitochondrial 21S rRNA, and roughly 100 in the rRNA of humans.
It has been demonstrated that pseudouridine in rRNA and tRNA fine-tunes and stabilizes the regional structure and aids in the maintenance of their roles in mRNA decoding, ribosome assembly, processing, and translation.
It has been demonstrated that pseudouridine in snRNA improves the interface between pre-mRNA and the spliceosomal RNA to help regulate splicing.
Fatty Acid Synthase
The FASN gene in humans encodes the enzyme known as fatty acid synthase (FAS). A multi-enzyme protein called fatty acid synthase catalyzes the synthesis of fatty acids.
It’s an entire enzymatic system, not just one enzyme, made up of two identical 272 kDa multifunctional polypeptides that transfer substrates from one functional domain to the next.
Its primary job is to use NADPH to catalyze the creation of palmitate (C16:0, a long-chain saturated fatty acid) from acetyl- and malonyl-CoA
Acetyl-CoA and malonyl-CoA are converted into fatty acids through a sequence of decarboxylative Claisen condensation processes.
Following each round of elongation, a ketoreductase (KR), dehydratase (DH), and enoyl reductase work in sequence to decrease the beta keto group to the completely saturated carbon chain (ER).
When the fatty acid chain has grown to a length of 16 carbons, it is released by the action of a thioesterase (TE), which is covalently connected to the phosphopantetheine prosthetic group of an acyl carrier protein (ACP) (palmitic acid).
Cellulose Synthase (UDP-Forming)
The primary enzyme responsible for producing cellulose is cellulose synthase (EC 2.4.1.12) in its UDP-forming form. It is generally referred to as UDP-glucose: (1→4) Enzymology’s 4-D-glucosyltransferase for D-glucan.
GDP-glucose is used by a related enzyme called cellulose synthase (GDP-forming) (EC 2.4.1.29). Both bacteria and plants have members of this family of enzymes.
Bacterial members may also be known as BcsA (bacterial cellulose synthase) or CelA, while plant members are typically known as CesA (cellulose synthase) or the speculative CslA (cellulose synthase-like) (simply “cellulose”).
CesA was acquired by plants as a result of the endosymbiosis that gave rise to the chloroplast. Family 2 of the glucosyltransferases include this one (GT2).
The majority of the biomass on Earth is produced through biosynthesis and hydrolysis by enzymes called glycosyltransferases.
The plant CesA superfamily is known to contain seven subfamilies, and the combined plant-algal superfamily contains 10.
The only animal group that possesses this enzyme is the urochordates, who obtained it through horizontal gene transfer more than 530 million years ago.
Cellulose Synthase (GDP-Forming)
This enzyme is a member of the hexosyltransferase subfamily of glycosyltransferases. This enzyme class is referred to by its scientific name, GDP-glucose:1,4-beta-D-glucan 4-beta-D-glucosyltransferase.
Other names that are frequently used are cellulose synthase (guanosine diphosphate-forming), cellulose synthetase, and guanosine diphosphoglucose-1,4-beta-glucan glucosyltransferase. This enzyme takes involvement in the metabolism of sucrose and starch.
What Is Synthetase?
The term “synthetase,” sometimes known as “ligase,” refers to any one of a class of roughly 50 enzymes that catalyzes chemical energy-conserving reactions and mediates between energy-consuming breakdown events and productive synthetic processes.
By cleaving an energetic phosphate bond, they generate the necessary energy to catalyze the combining of two molecules (in many cases, by the simultaneous conversion of adenosine triphosphate [ATP] to adenosine diphosphate [ADP]).
A ligase known as an amino acid-RNA ligase is one that catalyzes the creation of a carbon-oxygen bond between a transfer RNA and an amino acid.
When certain enzymes, such as amide synthetases, and peptide synthetases, are active, carbon-nitrogen (C-N) bonds are produced.
Difference Between Synthetase and Synthase
A synthetase is an enzyme that can catalyze the joining of two large molecules by creating a new chemical bond, typically with the simultaneous hydrolysis of a small pendant chemical group on one of the larger molecules, or it can catalyze the linking of two compounds, such as the joining of C-O, C-S, C-N, etc.
A ligase typically causes the following reaction to occur:
- A-C + b = Ab + C
- A+D + B + C + D + E + F = Ab + cD
Where the dependent, tiny groupings are represented by lowercase letters. Ligase may repair single-strand breaks that develop in double-stranded DNA during replication as well as link two complementary nucleic acid fragments.
On the other hand, synthase is an enzyme that catalyzes the synthesis process in biochemistry. They’re included in the category of lyases according to the EC number categorization.
Nomenclature
Keep in mind that the biological nomenclature initially distinguished between synthetases and synthases. In accordance with the original definition, synthetases utilize nucleoside triphosphates (such as ATP, GTP, CTP, TTP, and UTP) as a source of energy while synthases do not.
Nevertheless, according to the Joint Commission on Biochemical Nomenclature (JCBN), “synthase” can be used to refer to any enzyme that catalyzes synthesis (regardless of whether it employs nucleoside triphosphates), but “synthetase” should only be used to refer to “ligase.”
The primary distinction between Synthase and Synthetase is that Synthetase is a family of enzymes that may generate bonds between molecules, whereas Synthase is an enzyme.
| Synthase | Synthetase |
| Catalyzes a synthetic process without ATP | Requires ATP |
| Comes under vase or transferase classification | Comes under ligase classification |
| e.g. HMG-COA synthase, ATP synthase | e.g. Succiny1-COA synthetase, Glutamine synthetase |
Conclusion
- Synthetases don’t need NTPs to function since they catalyzed synthetic processes that required the hydrolysis of nucleoside triphosphates (not simply ATP).
- The International Union of Biochemistry’s Nomenclature Committee changed the definition of synthase in the 1980s to encompass all synthetic enzymes, whether or not they utilized NTPs, and synthetase became synonymous with ligase.
- Ligase is an enzyme that joins two smaller molecules together by utilizing the energy from NTP hydrolysis (usually through a condensation reaction).
Other Articles
- What Are the Differences Between Social Psychology and Cognitive Psychology? (Explained)
- Is 1ml of 200mg Testosterone Cypionate too Little to Make a Difference in Low Testosterone? (Facts)
- How Does A Pregnant Stomach Differ From A Fat Stomach? (Comparison)

